the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Plant functional traits determine latitudinal variations in soil microbial function: evidence from forests in China
Zhiwei Xu
Guirui Yu
Qiufeng Wang
Xinyu Zhang
Ruili Wang
Ning Zhao
Nianpeng He
Ziping Liu
Plant functional traits have increasingly been studied as determinants of ecosystem properties, especially for soil biogeochemical processes. While the relationships between biological community structures and ecological functions are a central issue in ecological theory, these relationships remain poorly understood at the large scale. We selected nine forests along the North–South Transect of Eastern China (NSTEC) to determine how plant functional traits influence the latitudinal pattern of soil microbial functions and how soil microbial communities and functions are linked at the regional scale. We found that there was considerable latitudinal variation in the profiles of different substrate use along the NSTEC. Specifically, we found that the substrate use by microorganisms was highest in the temperate forest soils (soil microbial substrate use intensities of 10–12), followed by the subtropical forest soils (soil microbial substrate use intensities of 7–10), and was least in the coniferous forest soils (soil microbial substrate use intensities of 4–7). The latitudinal variation in soil microbial function was more closely related to plant functional traits (leaf dry matter content, leaf C concentrations, and leaf N concentrations, P=0.002) than climate (mean annual precipitation, P=0.022). The soil silt, leaf dry matter, and leaf C and N contents were the main controls on the biogeographical patterns of microbial substrate use in these forest soils. The soil microbial community structures and functions were significantly correlated along the NSTEC. Soil carbohydrate and polymer substrate use were mainly related to soil Gram-positive (G+) bacterial and actinomycic phospholipid fatty acids (PLFAs), while the use of amine and miscellaneous substrates were related to soil Gram-negative (G−) bacterial and fungal PLFAs. The enzyme production varied with changes in the soil microbial communities. The soil enzyme activities were positively correlated with the bacterial PLFAs but were not correlated with the fungal PLFAs. The soil organic matter (SOM) decomposition rates were significantly higher in the temperate forests than in the subtropical and tropical forests, emphasizing the rapid degradability of high-energy substrates such as soil microbial biomass carbon, carbohydrates, and amino acids. The SOM decomposition rates were significantly and negatively related to soil dissolved organic carbon concentrations, carboxylic acids, polymers, and miscellaneous substrate use. The relationships between soil PLFAs and microbial substrate use, enzyme activities, and SOM decomposition rate show that as the soil microbial community structure changes, soil biogeochemical processes also change.
- Article
(2694 KB) -
Supplement
(463 KB) - BibTeX
- EndNote
The catabolic diversity of soil microbial communities is a useful indicator of how microbial functions adapt to environmental stress. It can be used to test fundamental questions about soil biological resistance and resilience (Jagadamma et al., 2014; Swallow and Quideau, 2015) and help us understand the role of microbial communities in different environments (Preston-Mafham et al., 2002). Biological community structure and function are intimately linked in ecological processes, and their relationships are a central issue in ecological theory (Talbot et al., 2014). Therefore, a major goal in ecological research is to identify and understand the mechanisms and relationships that control the structure and function of microbial communities over large spatial scales.
Numerous studies have documented how environmental and anthropogenic perturbations impact on the structure, diversity (Tu et al., 2016; Zhou et al., 2016), and enzyme activities (Peng and Wang, 2016; Xu et al., 2017) of soil microbial communities and have reported that forests in the same climatic zone develop similar microbial communities. Other researchers have examined spatial patterns in soil microbial function at different scales. For example, Tian et al. (2015), from their study of Changbai Mountain, China, found that the soil microbial metabolic activity and functional diversity were spatially dependent. Others reported that soil microbial activities varied by forest type, with high local variation and significant separation along regional climate gradients (Brockett et al., 2012; Cao et al., 2016). Soil microbes from different climatic zones have different affinities for carbon substrates. For example, microorganisms from boreal pine forest soils used carboxylic acids more efficiently but decomposed amino acids much less efficiently than microorganisms from temperate forest soils (Klimek et al., 2016). The soil microbial metabolic abilities are also influenced by the dominant tree species, through the production of chemically unique litter and root exudates, and the soil physicochemical properties (Menyailo et al., 2002). Despite this and because of limitations in analytical methods, questions still remain about how soil microbial functions vary at the regional scale.
The functional diversity of soil microbial communities is regulated by physicochemical soil properties (Gartzia-Bengoetxea et al., 2016), climate (Cao et al., 2016), and the composition of plant cover (Sherman and Steinberger, 2012). For example, the geographic patterns in soil microbial activities mainly reflect the climate, soil pH, and total phosphorus concentrations over large geographic scales (Cao et al., 2016). Research has shown that substrate-induced respiration rates were higher in soil microbial communities that developed under beech and holm oak forests than under oak and pine forests (Gartzia-Bengoetxea et al., 2016). Plant functional traits have increasingly been studied as determinants of ecosystem properties, especially for soil biogeochemical processes (De Vries et al., 2012; Pei et al., 2016). Soil bacteria phospholipid fatty acids (PLFAs) were found to be positively correlated with the community-weighted means (CWMs) of plant functional traits (leaf nitrogen (N) concentration) (De Vries et al., 2012). The plant leaf dry matter content and the leaf carbon (C) to nitrogen (N) ratio both influence the multivariate soil microbial community structure, and these factors positively promote the abundances of specific microbial functional groups (Pei et al., 2016). Limited soil resources, particularly in tropical forests, mean that soil microorganisms may be more reliant on plants than soil for C and nutrients via rhizosphere exudation or litter production, which varies among plant species (Russell et al., 2007; Raich et al., 2014; Waring et al., 2015). While soil functional diversity has been used as an indicator of microbial metabolic potential, there have been few studies of the integrated effects of climate, vegetation, and soil substrate availability on large-scale soil microbial functional diversity.
Although the functional characteristics of soil microorganisms are at least as important as their patterns of community structure in biogeochemical studies, the links between microbial community structure and microbial functions are poorly understood. There are two current hypotheses about how microbes determine ecosystem process rates. In functional redundancy, different microbes perform the same function and so changes in the microbial community structure do not necessarily lead to a change in soil function (Balser and Firestone, 2005; Strickland et al., 2009). For example, Banerjee et al. (2016) showed that the abundance of different bacterial and fungal groups changed by up to 300-fold under straw- and nutrient-amended treatments but that the decomposition rate remained similar, indicating possible functional redundancy. The functional redundancy hypothesis has recently been challenged by a counter-hypothesis, referred to as functional dissimilarity, which suggests that diversity brings stability and that every species plays a unique role in ecosystem function (Fierer and Bradford, 2007; Waldrop and Firestone, 2006). Soil microbial community composition therefore, combined with environmental variables, may ultimately determine ecosystem process rates. Waldrop and Firestone (2006) showed that Gram-positive bacteria (G+) were mainly responsible for the decomposition of pine needles and soil organic matter, but Gram-negative bacteria (G−) were mainly responsible for the decomposition of starch and xylose, which are easy to break down. Philippot et al. (2013), when studying the diversity of denitrifiers, showed that the loss of microbial diversity could result in decreases of between 4- and 5-fold in denitrification activity. In the Mediterranean, losses in the mass of decomposing leaf litter from shrub species accelerated as detritivore assemblages became more functionally dissimilar (Coulis et al., 2015). Research to date suggests that the different microbial communities will result in variations in soil microbial function and soil biochemical processes, so information about the relationships between soil microbial communities and their functions in natural ecosystems is urgently needed.
The North–South Transect of Eastern China (NSTEC) extends from a cold temperate coniferous forest in the north to a tropical rainforest in the south and includes almost all the forest types found in the Northern Hemisphere (Zhang and Yang, 1995) (Fig. 1 and Table 1). This transect, therefore, provides the optimal environment for investigating large-scale geographical patterns in microbial communities and their responses to environmental changes. In this study, we examined spatial patterns in soil labile C concentrations, soil organic matter (SOM) decomposition rates, and metabolic activity and functional diversity of microbes in nine forest biomes along the NSTEC. We assessed how abiotic factors, such as climate; soil physical and chemical properties; and biotic factors, in the form of community-weighted means (CWMs) of plant functional traits, contributed to soil functional diversity at the regional scale. We also examined the links between soil microbial community structure (PLFAs) and function (SOM decomposition rate, enzyme activities, and microbial substrate use). We tested four hypotheses in this study, as follows: (1) the profiles of soil microbial substrate use vary along a latitudinal gradient, (2) biogeographical patterns of soil microbial substrate use are constrained by climate and plant functional traits, and (3) different soil microbial communities may have substrate use profiles and SOM decomposition rates.
Table 1The main characteristics of the sampling sites along the North–South Transect of Eastern China.
a HZ, Huzhong; LS, Liangshui; CB, Changbai; DL, Dongling; TY, Taiyue; SN,
Shennong; JL, Jiulian; DH, Dinghu; JF, Jianfeng.
b MAT, mean annual temperature; MAP, mean annual precipitation.
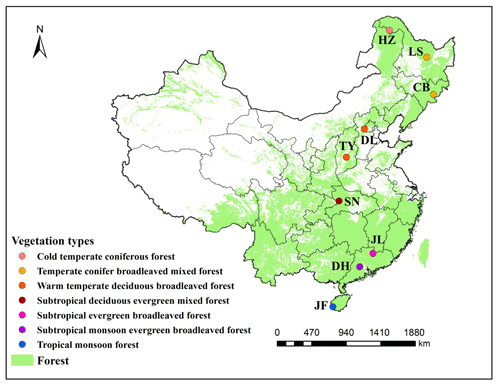
Figure 1Distribution of typical forest ecosystems along the North–South Transect of Eastern China (NSTEC). The abbreviations for the sampling sites from north to south are as follows: HZ, Huzhong; LS, Liangshui; CB, Changbai; DL, Dongling; TY, Taiyue; SN, Shennong; JL, Jiulian; DH, Dinghu; JF, Jianfeng. These abbreviations are used for the nine forests throughout.
2.1 Study area and soil sampling
We selected nine forest ecosystems along the NSTEC, namely Huzhong (HZ), Liangshui (LS), Changbai (CB), Dongling (DL), Taiyue (TY), Shennong (SN), Jiulian (JL), Dinghu (DH), and Jianfeng (JF) (18∘44′–51∘46′ N, 128∘53′–108∘51′ E) (Fig. 1, Table 1). Further information about the soil types and sites has been documented previously by Xu et al. (2017). Forest soils have been classified following the US soil taxonomy and are described in Table 1 (Soil Survey Staff, 2010), where information about the climate and the dominant vegetation at each site is also presented.
Soil samples were collected from four random plots at each site in July and August 2013, as described previously by Xu et al. (2017). Briefly, we established four sampling plots measured 30 m×40 m and collected soil samples from a depth of between 0 and 10 cm at between 30 and 50 points in each plot along an S-shape. On return to the laboratory, the fresh soil samples were immediately sieved through a 2 mm mesh and subdivided into three subsamples. One subsample was stored briefly at 4 ∘C until analysis for soil enzyme activities and soil pH. Another was stored briefly at −20 ∘C until analysis for PLFAs and Biolog EcoPlates. The third was air-dried, sieved through a 0.25 mm mesh, and analyzed for soil nutrients.
Table 2Soil properties of different sampling sites.
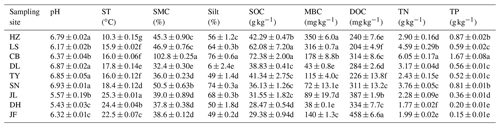
Note: ST: temperature of 0–10 cm soil; SMC: soil moisture content;
Silt: soil silt content; SOC: soil organic carbon; MBC: microbial biomass
carbon; DOC: dissolved organic carbon; TN: soil total nitrogen; TP: soil
total phosphorus. Values were presented as means±SE (n=4). Different lowercase letters indicate significant differences among forests.
The
abbreviations of the sampling sites were given in the Table 1.
2.2 Soil chemical analyses
Soil pH was measured at a soil:water ratio of 1:2.5. The soil moisture content (SMC) was measured gravimetrically on 20 g of fresh soil that was oven-dried at 105 ∘C to constant weight immediately on arrival at the study sites' laboratories (L. Liu et al., 2012). Soil organic carbon (SOC) and total N (TN) concentrations were determined by dry combustion of ground samples (100 mesh) in a C∕N analyzer (Elementar, Vario Max CN, Germany). Total phosphorus (TP) was determined with a flow injection autoanalyzer following digestion with H2SO4-HClO4 (Huang et al., 2011). After extraction with distilled water at a soil:distilled water ratio of 1:5, dissolved organic carbon (DOC) concentrations were determined by Liqui TOC II (Elementar, Liqui TOC II, Germany) (Jones and Willett, 2006). Soil microbial biomass carbon (MBC) was measured using the chloroform fumigation and direct extraction technique (Vance et al., 1987). A conversion factor of 2.64 was used to convert extracted C to biomass C. The silt fractions (<53 µm) of the samples were separated by wet-sieving and then were freeze-dried in the laboratory, as described by Six et al. (2000). The soil properties are shown in Table 2. We followed the method described by Bååth et al. (2003) for PLFA analysis, and PLFAs are expressed in units of nanomole per gram (nmol g−1). The four enzymatic activities of β-glucosidase (BG), N-acetylglucosaminidase (NAG), acid phosphatase (AP), and leucine aminopeptidase (LAP), responsible for soil C, N, and phosphorous cycling, were measured following the procedure outlined in Saiya-Cork et al. (2002) and are expressed in units of nanomole per hour per gram (). Information about PLFA and enzyme activities is presented in Table S1 in the Supplement.
The Biolog EcoPlates were purchased from Biolog, US. The substrates for BG, NAG, AP, and LAP were 4-methylumbelliferyl-β-D-glucoside, 4-methylumbelliferyl-N-acetyl-b-D-glucosaminide, 4-methylumbelliferyl-phosphate, and L-leucine-7-amino-4-methylcoumarin, respectively, and were stored at −20 ∘C. A methylumbelliferyl (MUB) standard was used for the BG, NAG, and AP enzymes, and an AMC (7-amino-4-methylcoumarin) standard was used for the LAP enzyme. The substrates and standards were purchased from Sigma. Analytical-grade reagents were used for the soil nutrient analysis.
2.3 Vegetation data
We established four sampling plots (30 m×40 m) in each forest ecosystem. In each plot, we recorded all the tree individuals and measured the height and diameter at breast height (DBH) of each woody individual with a DBH≥2 cm. The diversity of the tree species in the sampling plots was represented by H′, and the diversity (H′, Shannon–Wiener) of the tree species in the community was calculated as follows:
where Pi was the importance value of the species i as a proportion of all species and n was the number of the species.
We also calculated the community-weighted mean (CWM) values of the tree traits using the cover of each tree. As described by Xu et al. (2018), we collected litter and sun-exposed and mature leaves (leaf blades for trees) from between 5 and 10 individuals of each plant species at each site and determined their TN and total carbon (TC) concentrations. We calculated the specific leaf area (SLA, the one-sided area of a fresh leaf divided by its oven-dried mass, m2 kg−1), leaf dry matter content (LDMC, the oven-dried mass of a leaf divided by its water-saturated fresh mass, mg g−1), leaf C concentrations (leaf C, g kg−1), and leaf N concentrations (leaf N, g kg−1) for 10 fully expanded leaves of each sampled individual. To measure the leaf traits at the community level, we calculated the CWM of the tree layer as follows:
where pi is the relative contribution of the species i to the cover of the whole community, n is the number of the most abundant species, and traiti is the trait value of species i, as described by Garnier et al. (2004). The diversity of the tree species and plant functional traits are summarized in Table S2.
2.4 Microbial substrate use
Microbial functional diversities were determined using a Biolog EcoPlates™ (Biolog Inc., Hayward, California, USA) as described by Garland and Mills (1991). Briefly, approximately 10 g of fresh soil was suspended in 100 mL saline solution (0.85 % NaCl) and shaken on an orbital shaker for 30 min at 190 rpm. A 150 µL aliquot of supernatant from 1:1000 dilutions of each soil sample was added to each well. The plates were incubated at 25 ∘C, and the absorbance at 590 nm was measured using a microplate reader (GENios Pro™, Tecan Trading AG, Männedorf, Switzerland) every 24 up to 240 h (0, 24, 48, 72, 96, 20, 144, 168, 192, 216, and 240 h). To minimize the influence of cell density in comparisons among samples, results can be analyzed at constant average well color development (AWCD). The AWCD for each microplate was calculated by subtracting the control well optical density (OD) from the substrate well OD (blanked substrate wells), setting any resultant blanked substrate wells with negative values to 0, and taking the mean of the 95 blanked substrate wells.
The richness (R), Shannon–Wiener diversity index (H′), Shannon evenness index (E), and Simpson dominance index (D) were calculated from the absorption values after EcoPlate™ incubation for 96 h (Gomez et al., 2006). Additionally, the 31 C sources were divided into six groups, namely carbohydrates, carboxylic acids, amines, amino acids, polymers, and miscellaneous, as suggested by Zak et al. (1994). The average absorbance of all C sources within each group was computed as the intensity of the single substrate use. The soil total microbial metabolic intensities (S) of six carbon sources were estimated by the area underneath AWCD vs. t, and were obtained by integrating the equation against time t (Guckert et al., 1996):
where vi was the average optical density of the ith incubation time.
2.5 SOM decomposition rate
Four replicates from each sampling site with a 60 % water-holding capacity were incubated at 20 ∘C. In brief, 40 g of each fresh soil sample was put into a 150 mL incubation bottle, and the samples were then adjusted so that their moisture content corresponded to a water-holding capacity of 60 %. During the 4-week incubation period, the soil respiration rates were measured on days 1, 7, 14, 21, and 28 using an automatic system. The SOM decomposition rates were calculated as described in the study of Xu et al. (2015).
2.6 Statistical analysis
One-way analysis of variance (ANOVA) followed by a post hoc Tukey honestly significant difference test were used to test the significance of the differences among the soil properties, C use, functional diversity, and SOM decomposition rates in the different forest ecosystems. We tested the relationships between labile C, soil microbial community structure, microbial function, and the SOM decomposition rates with the Pearson correlation test. Differences were considered significant when P<0.05, with the marginal significance set at P<0.01. All P values were adjusted using the Bonferroni correction to account for multiple comparisons.
We used redundancy analysis (RDA) to examine the relationship between the environmental variables and soil microbial substrate use. The environmental variables were the same as those described in Xu et al. (2018), including climate, soil properties, litter properties, and plant functional traits. Before RDA, we conducted forward selection of the environmental variables that were significantly correlated with variations in the microbial substrate use profile using stepwise regression and the Monte Carlo permutation test. We used CANOCO software 4.5 (Ter Braak and Smilauer, 2000) for the RDA and stepwise regression. The environmental properties, which were significantly correlated with the microbial substrate use in the RDA, were stressed in the plots. Path analysis was conducted to examine the direct and indirect effect of biotic and abiotic factors on soil microbial use of carbon sources. All ANOVA, regression analyses, and path analysis were performed using SPSS 19.0 for Windows. Data are reported as the mean±SE (standard error).
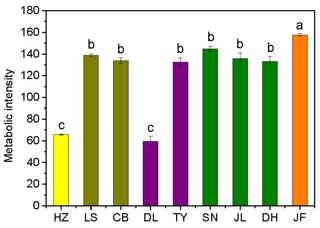
Figure 2Variations in soil microbial substrate use during a 240 h incubation for the nine forests. Different colors represent different forest types: yellow, coniferous forest; dark yellow, coniferous broad-leaved mixed forest; purple, deciduous broad-leaved forest; olive, subtropical evergreen broad-leaved forest; and orange, tropical monsoon forest. Different lowercase letters indicate significant differences among forests in the same climate zone. The abbreviations of the sampling sites are given in Table 1.
3.1 Patterns in the microbial substrate use, soil labile carbon, and SOM decomposition rates
There was no obvious latitudinal pattern for the soil total microbial metabolic intensity (Fig. 2). Of the forests along the NSTEC, the C metabolic intensity of soil microbes was lowest in HZ and LS; the C metabolic intensity of soil microbes differed significantly between JF and the other forests (Fig. 2), which indicates that the color development was significantly higher in the tropical forest soils than in the subtropical and temperate forest soils and is consistent with the variations in the AWCD (Fig. S1 in the Supplement). The average values of R, H′, and D were significantly different among the nine forest soils and were highest in JF, SN, and CB (Table 3). Generally, the soil microbial use of the six different carbon sources was relatively higher in tropical and subtropical climatic area (low latitude) than those in temperate climatic area (high latitude) (Fig. 3).
Table 3Functional diversity of soil microbial communities in forest ecosystems along the NSTEC.
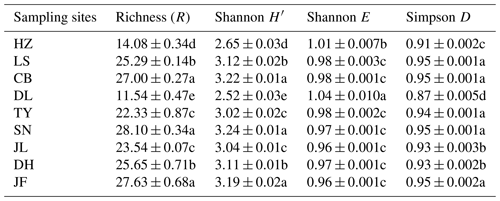
Indices were calculated based on the optical density values after incubation for 96 h. Data are expressed as means±standard errors. Different lowercase letters indicate significant differences among forests. The abbreviations of the sampling sites are the same as those used in Table 1.
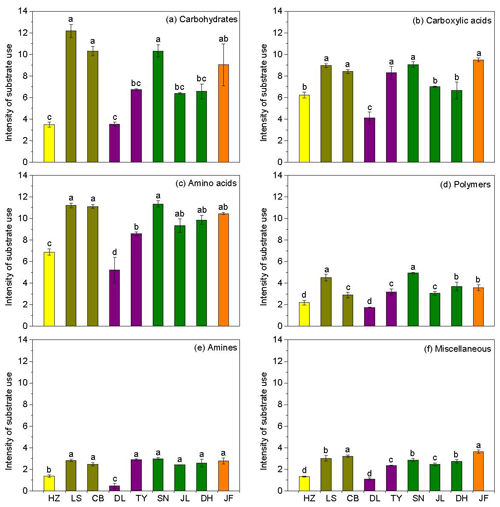
Figure 3Characteristics of microbial use of (a) carbohydrates, (b) carboxylic acids, (c) amino acids, (d) polymers, (e) amines, and (f) miscellaneous along the NSTEC. The representatives of different colors are shown in Fig. 2.
Across the nine forests, soil microorganisms used the six substrate groups in the same order; the carboxylic acid substrate was used most, followed by amino acids, carbohydrates, polymers, amines, and miscellaneous substrates (Fig. 3). Microorganisms in the boreal and temperate forests mainly metabolized carbohydrates, amino acids, and carboxylic acids, while those from the subtropical and tropical forests used the substrates in equal proportions. The substrate microbial use ability was highest in the coniferous broad-leaved mixed forest and tropical forest soils and lowest in the coniferous forest soil (Fig. 3).
Overall, soil MBC concentrations in the boreal and temperate forests were 3 to 8 times higher than those of the subtropical and tropical forests. In contrast, the average DOC concentrations in the tropical and subtropical forest soils, which ranged from 311 to 458 mg kg−1, were significantly higher than the average concentrations in the temperate and boreal forest soils, where the average concentrations ranged from 204 to 284 mg kg−1 (Table 2). The average SOM decomposition rates in the subtropical forests ranged from 0.64 to 2.42 and were significantly lower than the rates in the temperate forests, which ranged from 3.43 to 4.61 (Table S3).
3.2 Effect of environmental properties on soil microbial substrate use
Redundancy analysis showed that the variations in soil microbial substrate use were strongly and positively correlated with the CWM values of LDMC, leaf N, and leaf C and strongly and negatively correlated with the soil silt content and SMC (Fig. 4). The RDA2 of soil microbial substrate use was strongly positively correlated with TN and SOC, but negatively correlated with the mean annual precipitation (MAP) (Fig. 5). RDA1 mainly represented the plant functional traits, soil texture, and micrometeorological conditions, while RDA2 represented climate and soil nutrients. Overall, the soil silt content and the CWM values of plant functional traits were the main predictors of the latitudinal variation in the soil microbial substrate use along the NSTEC.
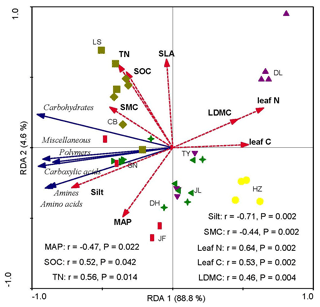
Figure 4Redundancy analysis (RDA) ordination biplot of soil microbial carbon sources use efficiency and environmental properties. The representatives of different colors are shown in Fig. 2. The dotted lines and solid lines represent the environmental variables and lipid signatures and carbon sources, respectively. The abbreviations of the variables in this figure are as follows: MAP, mean annual precipitation; LDMC, leaf dry matter weight; leaf C, leaf carbon content; leaf N, leaf nitrogen content; and SLA, specific leaf area. Soil properties included SMC, soil moisture content; Silt, soil silt content; TN, soil total nitrogen; and SOC, soil organic carbon. The abbreviations of the sampling sites are given in Table 1.

Figure 5The heatmap of the Pearson's correlation coefficients between the use of individual substrates and microbial PLFAs and soil enzyme activities. Note the abbreviations of the variables: actino-, actinomycetes; F∕B, fungi to bacteria ratio; G+, Gram-positive bacteria; G−, Gram-negative bacteria; , Gram-positive bacteria to Gram-negative bacteria ratio. BG, β-1, 4-glucosidase; NAG, β-1,4-N-acetylglucosaminidase; LAP, leucine aminopeptidase; and AP, acid phosphatase. ** P<0.01, * P<0.05.
3.3 Relationships between soil microbial substrate use, enzyme activities, and PLFAs
Microbial carbohydrate use was positively related to bacterial biomass and actinomycic biomass (Fig. 5). Microbial polymer use was negatively related with bacterial biomass and actinomycic biomass. Microbial amine use was negatively related to G− bacterial and fungal biomass. Miscellaneous substrate use was positively related to fungal biomass and bacterial biomass ratio (Fig. 5).
The abundance of G− bacteria was positively associated first with the specific activities of BG, whereas actinomycetes and G+ bacteria were positively associated with BG and LAP. Soil fungi were negatively associated with BG (Fig. 5).
3.4 Relationships between SOM decomposition rate, PLFAs, enzyme activity, and microbial metabolic activities
The SOM decomposition rates were significantly and positively related to soil MBC concentrations but significantly and negatively related to soil DOC concentrations (Fig. 6a and b). Except for amino acid and amine substrates, the SOM decomposition rates were significantly and positively related to microbial metabolic activities (AWCD) and carbohydrate substrate use (Fig. 6c and d) and negatively related to carboxylic acid, polymer, and miscellaneous substrate use (Fig. 6e, g, and i).
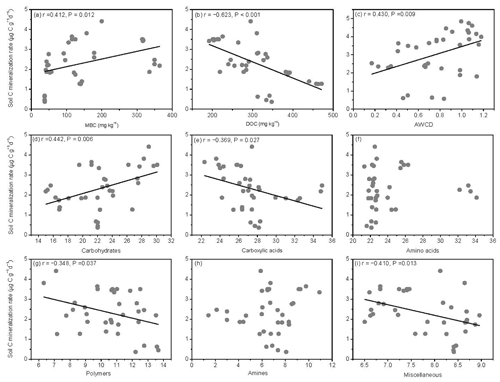
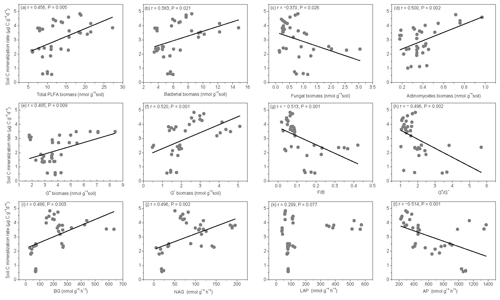
Figure 6Relationships between soil carbon mineralization rates () and microbial biomass C (MBC), soil dissolved organic C (DOC), average well color development (AWCD), and individual substrate use.
The SOM decomposition rates were significantly and positively correlated with total PLFAs (r=0.456, P=0.005), bacteria (r=0.3836, P=0.021), actinomycetes (r=0.500, P=0.002), and G− bacteria PLFAs (r=0.520, P=0.001) (Fig. 7a, b, d, and f) but were negatively correlated with fungal PLFAs (, P=0.026), F∕B (, P=0.001), and the (, P=0.002) (Fig. 7c, g, and h). Except for LAP activity, soil enzyme activities were significantly and positively correlated with the SOM decomposition rates (P<0.01) (Fig. 7i, j, and l).
4.1 Response of soil labile C and SOM decomposition rates to variations in forest type
Soil organic matter is one of the most important C pools in terrestrial ecosystems. The concentrations of soil DOC in the temperate forests were lower than those in subtropical forests, but the soil MBC concentrations were higher in temperate forests than in subtropical forests. This reflects the results of previous regional and global studies (Tian et al., 2010; Xu et al., 2013) and shows that the production to consumption ratio of soil DOC was lower but that microbial C immobilization was higher in the high-latitude forests than closer to the tropics (Fang et al., 2014). Soil DOC, as a labile SOM fraction with a rapid turnover, is one of the primary energy sources for microorganisms. The higher temperatures and precipitation in subtropical and tropical forests lead to higher turnover rates (Fang et al., 2014), so soil DOC concentrations were highest in subtropical forests and MBC concentrations were lowest in tropical forests. However, in temperate forests, more C is assimilated into microbial biomass so that less C is lost through chemical and physical processes (Liu et al., 2010). Also, because the decomposition ability of different microbe groups varies, the differences in soil microbial communities in different forest ecosystems may also be responsible for the spatial variations in the soil DOC and MBC concentrations along the NSTEC (Hagedorn et al., 2008).
Heterotrophic soil respiration is sustained by the decomposition of SOM. The SOM decomposition rates along the NSTEC were greater in temperate forests than in subtropical forests, which was consistent with the variations in the soil MBC and SOC concentrations. These results indicate that, as found in other studies, large-scale SOM decomposition rates are driven by the amounts of substrate available (Yu et al., 2010). Changes in the availability of C in SOM may affect the microbial resource strategies, which may in turn influence the SOM decomposition rate. Some forest soils were intermittently saturated (such as CB, Table 2) or high with mean annual precipitation. Under the anaerobic conditions, soil organic decomposition is mediated by a complex suite of microbial processes (Megonigal et al., 2004). The fermentation products including low-molecular-weight alcohols, fatty acids, and dihydrogen can serve as substrates for anaerobic respiration using a variety of alternative terminal electron acceptors in place of oxygen to mineralize organic carbon to carbon dioxide (CO2). Therefore, the soil organic matter decomposition rate might be slow in these anaerobic conditions. These results demonstrate that the reduction of organic matter is a key step of anaerobic decomposition (Keller and Takagi, 2013).
4.2 Mechanisms driving latitudinal variations in microbial substrate use
The AWCD reflects the sole C source use ability of the soil microbial community (Garland and Mills, 1991). Of the six groups of C substrates, microbial communities in the temperate forests mainly used carbohydrates, carboxylic acids, and amino acids, which suggests that microorganisms in temperate forests probably use high-energy substrates that degrade easily (Kunito et al., 2009). The carbon substrate use was lowest in the boreal coniferous forest (HZ). This shows that, compared with coniferous species, broad-leaved tree species produce root exudates and litter high in water-soluble sugars, organic acids, and amino acids that are more favorable for microbial activity (Priha et al., 2001).
There was no significant latitudinal pattern in the soil total microbial metabolic intensity (S) in our study, which was inconsistent with hypothesis 1. However, there was significant latitudinal variation in the use of the six different carbon sources. The soil microbial use of carbon sources was relatively higher in tropical and subtropical climatic area than those in temperate climatic areas. Consistent with hypothesis 2, soil microbial functions were similar and closely related to the CWM values of the tree species (Fig. 4). However, only MAP of climatic factors had a moderate effect on the soil microbial function (Fig. 4). Climate may have an indirect effect on the latitudinal pattern of different carbon sources use. In our study, MAT and MAP affected the six different carbon sources use indirectly by influencing the soil temperature (ST), soil total nitrogen (TN), and total phosphorus (TP) (Tables S4–S9). It was reported that climate was the main environmental parameter driving the latitudinal patterns of community-level leaf traits through the regulation of species composition (R. Wang et al., 2016). Woody plant species with evergreen broadleaves dominated in the tropic regions characterized by hot, humid, and infertile habitats showed low CWM values of leaf N and SLA and high CWM values of LDMC (Table S2).
A growing number of studies reported that vegetation type, land use, soil nutrients, and soil organic matter quality and quantity can determine large-scale patterns of microbial communities (de Vries et al., 2012; Tu et al., 2016). Plant functional traits that are related to growth may determine a tree species' ability to contribute to the soil carbon pool via leaf litter inputs. For example, it was previously reported that plant traits such as the leaf N content, SLA, and LDMC could explain variations in soil nutrients and litter decomposition rates (Eichenberg et al., 2014; Laughlin, 2011). Therefore, we examined how these plant traits influenced the soil microbial function by latitude. We found that changes in the soil microbial C substrate use with latitude were mainly related to the soil silt contents and the CWMs of LDMC, and leaf C and leaf N concentrations, which indicates that the quality of nutrients from plants had a major influence on microbial carbon use efficiency (hypothesis 2). Plant species with high SLA, high leaf N concentrations, and low LDMC can produce bacteria-dominated soil microbial communities in grasslands (Orwin et al., 2010). Looking beyond individual traits, related tree species may cultivate microbial communities with similar preferences for carbon sources through coevolution of plants and microbes (X. Liu et al., 2012; Buscot, 2015).
As hypothesized, the soil microbial community composition was explained by the CWMs of plant traits at the regional scale. Carbon substrate use was negatively correlated with the CWM of leaf N concentrations (Table S2, Fig. S2). Bacterially dominated soil microbial communities develop from leaf litter comprised of N-rich leaves from fast-growing species (De Vries et al., 2012), while leaves with low N concentrations will promote fungal domination (Orwin et al., 2010; De Vries et al., 2012). In line with this, fungal biomass decreased, and bacterial biomass increased, as the CWM leaf N content increased and is associated with fast-growing N-exploitative plants (Xu et al., 2018). Leaf N concentrations are considered indicators of plant growth and resource uptake (Wright et al., 2004). The results from this study show that, along the NSTEC, high leaf N restrained microbial C substrate use (Fig. S2) and was a good indicator of the competition between plants for soil N (Pei et al., 2016). Soil microbes and nearby plants may have been competing for N in the soil.
We also found that the C substrate use was negatively correlated with the CWM of leaf C concentrations (Table S2, Fig. S2). Plants at high latitudes may have higher leaf C concentrations than plants at lower latitudes so that they can balance the osmotic pressure of cells and resist freezing (Millard et al., 2007). The increased C was most likely in the form of an increase in nonstructural C, including starch, low-molecular-weight sugars, and storage lipids that are easy to break down. Therefore, soil microorganisms from the temperate forests mainly metabolized high-energy substrates such as carbohydrates, carboxylic acids, and amino acids.
The LDMC is the ratio of the leaf dry matter weight to the fresh weight and has been used as a proxy for the ratio of structural compounds to assimilatory tissue (mesophyll and epidermis; Van Arendonk and Poorter, 1994). High values of LDMC indicate large amounts of vascular tissue, cellulose, insoluble sugars and leaf lignin that are difficult to decompose (Poorter and Bergkotte, 1992); C substrates such as carbohydrates, carboxylic acid, and amino acid are, however, easy to decompose (Myers et al., 2001). In line with this, the use of carbohydrate, carboxylic acid, and amino acid substrates was negatively related to the CWMs of the LDMC (Table S2). Pei et al. (2016) reported that the LDMC was an important driver of multivariate soil microbial community structure and G− bacterial abundance.
Soil texture regulates soil biological processes and so affects the soil microbial community structure (Sessitsch et al., 2001). In the present study, microbial C substrate use was significantly and positively related to the soil silt content. Soil types and textures varied along the NSTEC. Soil texture influences how microbes use organic matter and has a strong influence on soil moisture, nutrient availability, and retention (Veen and Kuikman, 1990). Fine-textured soils with a higher silt content are known to be more favorable for bacterial growth than soils with a lower silt content because of their greater water-holding capacity and nutrient availability, and because they are better protected from bacterial grazers (Carson et al., 2010). We found that the microbial C substrate use was higher in LS, CB, SN, and JL than in the other forests, reflecting their fine-grained soils and high silt contents, which ranged from 60 % to 80 %.
4.3 Links between soil microbial community structure and function
The soil microbial community structure and functions were significantly correlated along the NSTEC. Soil carbohydrate and polymer substrate use were mainly related to soil G+ bacterial and actinomycic biomass, but amines and miscellaneous substrates were mainly related to soil G− bacterial biomass, fungal biomass, and the F∕B ratio. Soil bacteria mainly decomposed simple carbohydrates, organic acids, and amino acids, whereas soil fungi mainly decomposed recalcitrant compounds (Myers et al., 2001; Treonis et al., 2004). Consistent with this, soil bacterial PLFAs were positively correlated with the carbohydrate substrate use and the fungal PLFAs were positively related to miscellaneous substrate use. The results are similar to those reported by Sterner and Elser (2002), who found that fungi tended to have higher C∕N or C∕P ratios while heterotrophic bacteria typically have lower C∕N or C∕P ratios.
Shifts in microbial community composition may influence enzyme production (DeForest et al., 2012; Waldrop et al., 2000; Brockett et al., 2012). Different microbial groups require different amounts of nutrients to construct biomass or have enzymes that differ in their affinity for nutrients. We found that the relative abundances of the G+ bacteria and actinomycetes communities were associated with the specific activities of BG, AP, and LAP, whereas the relative abundance of the G− bacteria was correlated with soil NAG activities involved in chitin degradation. In agreement with our study, numerous other researchers have reported significant correlations between PLFA profiles and enzyme activities (Waldrop et al., 2000; DeForest et al., 2012; Brockett et al., 2012; Riah-Anglet et al., 2015). Soil BG was mainly responsible for cellulose degradation and was involved in breaking down complex organic compounds (cellobiose) into small-molecule substrates (glucose) in favor of acquiring C through microbial community growth (Waldrop et al., 2000). Soil NAG activities were weakly and positively related to fungal biomass in the present study and may have been mainly produced by fungal populations (Valášková et al., 2007). Fungi are commonly considered producers of oxidative enzymes. Therefore, the influence of fungal biomass on variations in enzyme activities was minimal (Kivlin and Treseder, 2014). The linkages between enzyme activity and community composition may provide some insight into the microbial mechanisms that drive the decomposition of macromolecular C compounds. These results suggest that overall ecosystem functioning may suffer if soil microbial groups are lost.
The quality and amounts of SOM are influenced by the biomass, vegetation coverage, root distribution, and microbial species (Raich and Schlesinger, 1992). The SOM decomposition rates were higher in temperate forests than in tropical forests and may reflect the higher soil microbial biomass (Q. Wang et al., 2016). In line with this, SOM decomposition rates were positively related to soil MBC concentrations and different groups of PLFAs. The inverse relationships between SOM decomposition rates and DOC, carboxylic acids, polymers, and miscellaneous along the NSTEC, indicate a shift in the soil C turnover from open to closed with increases in the soil labile C concentrations (Fang et al., 2014). Soil DOC and MBC influence SOM decomposition rates indirectly by regulating microbial properties (Boberg et al., 2014; Wei et al., 2014). In our study, SOM decomposition rates were positively related to bacterial PLFAs but negatively with fungal PLFAs. Because different communities of microbes have different SOM use efficiencies (Balser and Wixon, 2009; Lipson et al., 2009; Monson et al., 2006), changes in the microbial community structure may influence the microbial activities and the decomposition rates of organic matter (Lipson et al., 2009; Keiblinger et al., 2010). The functional dissimilarity of microbes and fungi may help explain these results. However, we did not measure some key variables, such as the microbial competition and interactions, and relationship between the microbial diversity and the decomposition rates. Therefore, in the future, we will use different experimental techniques that will help us gain an improved understanding of the mechanisms that drive the relationships between the structure and function of microbial communities.
In this study, we examined the patterns in labile C concentrations, SOM decomposition rates, microbial substrate use, and functional diversity and identified a combination of abiotic and biotic factors that influenced soil microbial functional diversity at the regional scale. The MBC concentration and SOM decomposition rates were significantly lower and the soil DOC concentrations were higher in the subtropical and tropical forests than in the temperate forests. There was no obvious latitudinal pattern for the soil total microbial metabolic intensity. However, soil microbial use of the different carbon sources varied with the latitude except the amines carbon source. Soil microbial use of carbon sources was relatively higher in tropical climatic areas. For the first time, we showed that CWM values of plant traits explained variations in soil microbial C substrate use at the regional scale. Additionally, the fine-grained soils with high silt contents were higher in the microbial C substrate use. Climate factors affected the soil microbial uses of carbon sources indirectly by influencing the soil temperature (ST) and soil nutrition. Soil microbial community structure and function were strongly related, which suggests that the loss of soil microbial groups may have consequences for overall ecosystem functioning.
Requests for data and materials should be addressed to Nianpeng He (henp@igsnrr.ac.cn) and Guirui Yu (yugr@igsnrr.ac.cn).
The supplement related to this article is available online at: https://doi.org/10.5194/bg-16-3333-2019-supplement.
ZX, GY, and XZ planned and designed the research. ZX, NH, RW, and NZ conducted fieldwork. ZX, GY, XZ, and QW wrote the article. All authors contributed critically to the drafts and gave final approval for publication.
The authors declare that they have no conflict of interest.
This research was jointly supported by the National Natural Science Foundation of China (41601084 and 41571251), the Fundamental Research Funds for the Central Universities (2412019FZ001), Science and Technology Research Project of Jilin Province (JJKH20190283KJ), the National Key R&D Program of China (2016YFA0602301), and the China Postdoctoral Science Foundation (2018M631850).
This research has been supported by the National Natural Science Foundation of China (41601084 and 41571251), the Fundamental Research Funds for the Central Universities (2412019FZ001), Science and Technology Research Project of Jilin Province (JJKH20190283KJ), the National Key R&D Program of China (2016YFA0602301), and the China Postdoctoral Science Foundation (2018M631850).
This paper was edited by Yakov Kuzyakov and reviewed by four anonymous referees.
Bååth, E. and Anderson, T. H.: Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques, Soil Biol. Biochem., 35, 955–963, 2003.
Balser, T. C. and Firestone, M. K.: Linking microbial community composition and soil processes in a California annual grassland and mixed-conifer forest, Biogeochemistry, 73, 395–415, 2005.
Balser, T. C. and Wixon, D. L.: Investigating biological control over soil carbon temperature sensitivity, Global Change Biol., 15, 2935–2949, 2009.
Banerjee, S., Kirkby, C. A., Schmutter, D., Bissett, A., Kirkegaard, J. A., and Richardson, A. E.: Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil, Soil Biol. Biochem., 97, 188–198, 2016.
Boberg, J. B., Finlay, R. D., Stenlid, J., Ekblad, A., and Lindahl, B. D.: Nitrogen and carbon reallocation in fungal mycelia during decomposition of boreal forest litter, PloS one, 9, e92897, https://doi.org/10.1371/journal.pone.0092897, 2014.
Brockett, B. F. T., Prescott, C. E., and Grayston, S. J.: Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada, Soil Biol. Biochem., 44, 9–20, 2012.
Buscot, F.: Implication of evolution and diversity in arbuscular and ectomycorrhizal symbioses, J. Plant Physiol., 172, 55–61, 2015.
Cao, H., Chen, R., Wang, L., Jiang, L., Yang, F., Zheng, S., Wang, G., and Lin, X.: Soil pH, total phosphorus, climate and distance are the major factors influencing microbial activity at a regional spatial scale, Sci. Rep.-UK, 6, 25815, https://doi.org/10.1038/srep25815, 2016.
Carson, J. K., Gonzalez-Quinones, V., Murphy, D. V., Hinz, C., Shaw, J. A., and Gleeson, D. B.: Low pore connectivity increases bacterial diversity in soil, Appl. Environ. Microb., 76, 3936–3942, 2010.
Coulis, M., Fromin, N., David, J.-F., Gavinet, J., Clet, A., Devidal, S., Roy, J., and Hättenschwiler, S.: Functional dissimilarity across trophic levels as a driver of soil processes in a Mediterranean decomposer system exposed to two moisture levels, Oikos, 124, 1304–1316, 2015..
DeForest, J., Smemo, K., Burke, D., Elliott, H., and Becker, J.: Soil microbial responses to elevated phosphorus and pH in acidic temperate deciduous forests, Biogeochemistry, 109, 189–202, 2012.
De Vries, F. T., Manning, P., Tallowin, J. R., Mortimer, S. R., Pilgrim, E. S., Harrison, K. A., Hobbs, P. J., Quirk, H., Shipley, B., Cornelissen, J. H., Kattge, J., and Bardgett, R. D.: Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities, Ecol. Lett., 15, 1230–1239, 2012.
Eichenberg, D., Trogisch, S., Huang, Y., He, J. S., and Bruelheide, H.: Shifts in community leaf functional traits are related to litter decomposition along a secondary forest succession series in subtropical China, J. Plant Ecol., 8, 401–410, https://doi.org/10.1093/jpe/rtu021, 2014.
Fang, H., Cheng, S., Wang, Y., Yu, G., Xu, M., Dang, X., Li, L., and Wang, L.: Changes in soil heterotrophic respiration, carbon availability, and microbial function in seven forests along a climate gradient, Ecol. Res., 29, 1077–1086, 2014.
Fierer, N. and Bradford, M. A.: Toward an ecological classification of soil bacteria, Ecology, 88, 1354–1364, 2007.
Garland, J. L. and Mills, A. L.: Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole carbon source utilization, Appl. Environ. Microb., 57, 2351–2359, 1991.
Garnier, E., Cortez, J., Billes, G., Navas, M. L., Roumet, C., Debussche, M., Gérard, L., Alain, B., David, A., Astrid, B., Cathy, N., and Jean-Patrick, T.: Plant functional markers capture ecosystem properties during secondary succession, Ecology, 85, 2630–2637, 2004.
Gartzia-Bengoetxea, N., Kandeler, E., Martínez de Arano, I., and Arias-González, A.: Soil microbial functional activity is governed by a combination of tree species composition and soil properties in temperate forests, Appl. Soil Ecol., 100, 57–64, 2016.
Gomez, E., Ferreras, L., and Toresani, S.: Soil bacterial functional diversity as influenced by organic amendment application, Bioresour. Technol., 97, 1484–1489, 2006.
Guckert, J. B., Carr, G. J., Johnson, T. D., Hamm, B. G., Davidson, D. H., and Kumagai, Y.: Community analysis by Biolog: curve integration for statistical analysis of activated sludge microbial habitats, J. Microbiol. Meth., 27, 183–197, 1996.
Hagedorn, F., van Hees, P. A., Handa, I. T., and Hättenschwiler, S.: Elevated atmospheric CO2 fuels leaching of old dissolved organic matter at the alpine treeline, Global Biogeochem. Cy., 22, https://doi.org/10.1029/2007GB003026, 2008.
Huang, Z., Clinton, P. W., Baisden, W. T., and Davis, M. R.: Long-term nitrogen additions increased surface soil carbon concentration in a forest plantation despite elevated decomposition, Soil Biol. Biochem., 43, 302–307, 2011.
Jagadamma, S., Mayes, M. A., Steinweg, J. M., and Schaeffer, S. M.: Substrate quality alters the microbial mineralization of added substrate and soil organic carbon, Biogeosciences, 11, 4665–4678, https://doi.org/10.5194/bg-11-4665-2014, 2014.
Jones, D. and Willett, V.: Experimental evaluation of methods to quantify dissolved organic nitrogen (DON) and dissolved organic carbon (DOC) in soil, Soil Biol. Biochem., 38, 991–999, 2006.
Keiblinger, K. M., Hall, E. K., Wanek, W., Szukics, U., Hammerle, I., Ellersdorfer, G., Bock, S., Strauss, J., Sterflinger, K., Richter, A., and Zechmeister-Boltenstern, S.: The effect of resource quantity and resource stoichiometry on microbial carbon-use-efficiency, FEMS Microb. Ecol., 73, 430–440, 2010.
Keller, J. K. and Takagi, K. K.: Solid-phase organic matter reduction regulates anaerobic decomposition in bog soil, Ecosphere, 4, 1–12, 2013.
Kivlin, S. N. and Treseder, K. K.: Soil extracellular enzyme activities correspond with abiotic factors more than fungal community composition, Biogeochemistry, 117, 23–37, 2014.
Klimek, B., Chodak, M., Jaźwa, M., and Niklińska, M.: Functional diversity of soil microbial communities in boreal and temperate Scots pine forests, Eur. J. Forest Res., 135, 731–742, 2016.
Kunito, T., Akagi, Y., Park, H.-D., and Toda, H.: Influences of nitrogen and phosphorus addition on polyphenol oxidase activity in a forested Andisol, Eur. J. Forest Res., 128, 361–366, 2009.
Laughlin, D. C.: Nitrification is linked to dominant leaf traits rather than functional diversity, J. Ecol., 99, 1091–1099, 2011.
Lipson, D. A., Monson, R. K., Schmidt, S. K., and Weintraub, M. N.: The trade-off between growth rate and yield in microbial communities and the consequences for under-snow soil respiration in a high elevation coniferous forest, Biogeochemistry, 95, 23–35, 2009.
Liu, L., Gundersen, P., Zhang, T., and Mo, J. M.: Effects of phosphorus addition on soil microbial biomass and community composition in three forest types in tropical China, Soil Biol. Biochem., 44, 31–38, 2012.
Liu, X., Liang, M., Etienne, R.S., Wang, Y., Staehelin, C., and Yu, S.: Experimental evidence for a phylogenetic Janzen-Connell effect in a subtropical forest, Ecol. Lett., 15, 111–118, 2012.
Liu, Z., Liu, G., Fu, B., Wu, Y., Hu, H., and Fu, S.: Changes in the soil microbial community with a pine plantation restoration in a dry valley of the upper reaches of the Minjiang River, southwest China, Ann. N. Y. Acad. Sci., 1195, E82–95, 2010.
Megonigal, J. P., Hines, M. E., and Visscher, P. T.: Anaerobic metabolism: linkages to trace gases and aerobic processes, in: Biogeochemistry, edited by: Schlesinger, W. H., pp. 317–424, Elsevier-Pergamon, Oxford, UK, 2004.
Menyailo, O. V., Hungate, B. A., and Zech, W.: Tree species mediated soil chemical changes in a Siberian artificial Afforestation experiment, Plant Soil, 242, 171–182, 2002.
Millard, P., Sommerkorn, M., and Grelet, G. A.: Environmental change and carbon limitation in trees: a biochemical, ecophysiological and ecosystem appraisal, New Phytol., 175, 11–28, 2007.
Monson, R. K., Lipson, D. L., Burns, S. P., Turnipseed, A. A., Delany, A. C., Williams, M. W., and Schmidt, S. K.: Winter forest soil respiration controlled by climate and microbial community composition, Nature, 439, 711–714, 2006.
Myers, R. T., Zak, D. R., White, D. C., and Peacock, A.: Landscape-level patterns of microbial community composition and substrate use in upland forest ecosystems, Soil Sci. Soc. Am. J., 65, 359–367, 2001.
Orwin, K. H., Buckland, S. M., Johnson, D., Turner, B. L., Smart, S., Oakley, S., and Bardgett, R. D.: Linkages of plant traits to soil properties and the functioning of temperate grassland, J. Ecol., 98, 1074–1083, 2010.
Pei, Z., Eichenberg, D., Bruelheide, H., Kröber, W., Kühn, P., Li, Y., von Oheimb, G., Purschke, O., Scholten, T., Buscot, F., and Gutknecht, J. L. M.: Soil and tree species traits both shape soil microbial communities during early growth of Chinese subtropical forests, Soil Biol. Biochem., 96, 180–190, 2016.
Peng, X. and Wang, W.: Stoichiometry of soil extracellular enzyme activity along a climatic transect in temperate grasslands of northern China, Soil Biol. Biochem., 98, 74–84, 2016.
Philippot, L., Spor, A., Henault, C., Bru, D., Bizouard, F., Jones, C. M., Sarr, A., and Maron, P. A.: Loss in microbial diversity affects nitrogen cycling in soil, ISME J., 7, 1609–1619, 2013.
Poorter, H. and Bergkotte, M.: Chemical composition of 24 wild species differing in relative growth rate, Plant Cell Environ., 15, 221–229, 1992.
Preston-Mafham, J., Boddy, L., and Randerson, P. F.: Analysis of microbial community functional diversity using sole-carbon-source utilisation profies-a critique, FEMS Microb. Ecol., 42, 1–14, 2002.
Priha, O., Grayston, S. J., Hiukka, R., Pennanen, T., and Smolander, A.: Microbial community structure and characteristics of the organic matter in soils under Pinus sylvestris, Picea abies and Betula pendula at two forest sites, Biol. Fertil. Soils, 33, 17–24, 2001.
Raich, J. W. and Schlesinger, W. H.: The global carbon dioxide flux in soil respiration and its relationship to vegetation and climate, Tellus B, 44, 81–99, 1992.
Raich, J. W., Clark, D. A., Schwendenmann, L., and Wood, T. E.: Aboveground tree growth varies with belowground carbon allocation in a tropical rainforest environment, PloS one, 9, e100275, https://doi.org/10.1371/journal.pone.0100275, 2014.
Riah-Anglet, W., Trinsoutrot-Gattin, I., Martin-Laurent, F., Laroche-Ajzenberg, E., Norini, M.-P., Latour, X., and Laval, K.: Soil microbial community structure and function relationships: A heat stress experiment, Appl. Soil Ecol., 86, 121–130, 2015..
Russell, A. E., Raich, J. W., Valverde-Barrantes, O. J., and Fisher, R. F.: Tree species effects on soil properties in experimental plantations in tropical moist forest, Soil Sci. Soc. Am. J., 71, 1389–1397, 2007.
Saiya-Cork, K. R., Sinsabaugh, R. L., and Zakb, D. R.: The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil, Soil Biol. Biochem., 34, 1309–1315, 2002.
Sessitsch, A., Weilharter, A., Gerzabek, M. H., Kirchmann, H., and Kandeler, E.: Microbial Population Structures in Soil Particle Size Fractions of a Long-Term Fertilizer Field Experiment, Appl. Environ. Microb., 67, 4215–4224, 2001.
Sherman, C. and Steinberger, Y.: Microbial functional diversity associated with plant litter decomposition along a climatic gradient, Microb. Ecol., 64, 399–415, 2012.
Six, J., Elliott, E. T., and Paustian, K.: Soil structure and soil organic matter: II. A normalized stability index and the effect of mineralogy, Soil Sci. Soc. Am. J., 64, 1042–1049, 2000.
Soil Survey Staff: Keys to Soil Taxonomy, 11th edn., USDA-Natural Resources Conservation Service, Washington, D.C., 2010.
Sterner, R. W. and Elser, J. J.: Ecological stoichiometry: the biology of elements from molecules to the biosphere, Princeton University Press, Princeton, New Jersey, USA, 2002.
Strickland, M. S., Lauber, C., Fierer, N., and Bradford, M. A.: Testing the functional significance of microbial community composition, Ecology, 90, 441–451, 2009.
Swallow, M. J. B. and Quideau, S. A.: A method for determining community level physiological profiles of organic soil horizons, Soil Sci. Soc. Am. J., 79, 536–542, 2015.
Talbot, J. M., Bruns, T. D., Taylor, J. W., Smith, D. P., Branco, S., Glassman, S. I., Erlandson, S., Vilgalys, R., Liao, H. L., Smith, M. E., and Peay, K. G.: Endemism and functional convergence across the North American soil mycobiome, P. Natl. Acad. Sci. USA, 111, 6341–6346, 2014.
Ter Braak, C. J. F. and Smilauer, P.: CANOCO Reference manual and CanoDraw for Windows User's guide: Software for Canonical Community Ordination (Version 4.5), Microcomputer Power, Ithaca, NY, 2000.
Tian, H., Chen, G., Zhang, C., Melillo, J. M., and Hall, C. A. S.: Pattern and variation of ratios in China's soils: a synthesis of observational data, Biogeochemistry, 98, 139–151 2010.
Tian, J., McCormack, L., Wang, J., Guo, D., Wang, Q., Zhang, X., Yu, G., Blagodatskaya, E., and Kuzyakov, Y.: Linkages between the soil organic matter fractions and the microbial metabolic functional diversity within a broad-leaved Korean pine forest, Eur. J. Soil Biol., 66, 57–64, 2015.
Treonis, A. M., Ostle, N. J., Stott, A. W., Primrose, R., Grayston, S. J., and Ineson, P.: Identification of groups of metabolically-active rhizosphere microorganisms by stable isotope probing of PLFAs, Soil Biol. Biochem., 36, 533–537, 2004.
Tu, Q., Deng, Y., Yan, Q., Shen, L., Lin, L., He, Z., Wu, L., Van Nostrand, J. D., Buzzard, V., Michaletz, S. T., Enquist, B. J., Weiser, M. D., Kaspari, M., Waide, R. B., Brown, J. H., and Zhou, J.: Biogeographic patterns of soil diazotrophic communities across six forests in the North America, Mol. Ecol., 25, 2937–2948, 2016.
Valášková, V., Šnajdr, J., Bittner, B., Cajthaml, T., Merhautová, V., Hofrichter, M., and Baldrian, P.: Production of lignocellulose-degrading enzymes and degradation of leaf litter by saprotrophic basidiomycetes isolated from a Quercus petraea forest, Soil Biol. Biochem., 39, 2651–2660, 2007.
van Arendonk, J. J. C. M. and Poorter, H.: The chemical composition and anatomical structure of leaves of grass species differing in relative growth rate, Plant Cell Environ., 17, 963–970, 1994.
Vance, E. D., Brookes, P. C., and Jenkinson, D. S.: An extraction method for measuring soil microbial biomass C, Soil Biol. Biochem., 19, 703–707, 1987.
Veen, J. A. V. and Kuikman, P. J.: Soil structural aspects of decomposition of organic-matter by micro-organism, Biogeochemistry, 11, 213–233, 1990.
Waldrop, M. P. and Firestone, M. K.: Response of microbial community composition and function to soil climate change, Microb. Ecol., 52, 716–724, 2006.
Waldrop, M. P., Balser, T. C., and Firestone, M. K.: Linking microbial community composition to function in a tropical soil, Soil Biol. Biochem., 32, 1837–1846, 2000.
Wang, Q., He, N., Yu, G., Gao, Y., Wen, X., Wang, R., Koerner, S. E., and Yu, Q.: Soil microbial respiration rate and temperature sensitivity along a north-south forest transect in eastern China: Patterns and influencing factors, J. Geophys. Res.-Biogeo., 121, 399–410, 2016.
Wang, R., Yu, G., He, N., Wang, Q., Zhao, N., and Xu, Z.: Latitudinal variation of leaf morphological traits from species to communities along a forest transect in eastern China, J. Geogr. Sci., 26, 15–26, 2016.
Waring, B. G., Alvarez-Cansino, L., Barry, K. E., Becklund, K. K., Dale, S., Gei, M. G., Keller, A. B., Lopez, O. R., Markesteijn, L., Mangan, S., Riggs, C. E., Rodriguez-Ronderos, M. E., Segnitz, R. M., Schnitzer, S. A., and Powers, J. S.: Pervasive and strong effects of plants on soil chemistry: a meta-analysis of individual plant `Zinke' effects, Proc. R. Soc. B. The Royal Society, 282, 20151001, https://doi.org/10.1098/rspb.2015.1001, 2015.
Wei, H., Guenet, B., Vicca, S., Nunan, N., AbdElgawad, H., Pouteau, V., Shen, W., and Janssens, I. A.: Thermal acclimation of organic matter decomposition in an artificial forest soil is related to shifts in microbial community structure, Soil Biol. Biochem., 71, 1–12, 2014.
Wright, I. J., Reich, P. B., Westoby, M., Ackerly, D. D., Baruch, Z., Bongers, F., Cavender-Bares, J., Chapin, T., Cornelissen, J. H. C., Diemer, M., Flexas, J., Gaenier, E., Geoom, P. K., Gulias, J., Hikosaka, K., Lamont, B. B., Lee, T., Lee, W., Lusk, C., Midgley, J. J., Navas, M.-L., Niinemets, Ü., Oleksyn, J., Osada,N., Poorter, H., Poot, P., Prior, L., Pyankov, V. I., Roumet, C., Thomas, S. C., Tjoelker, M. G., Veneklaas, E. J., and Villar, R.: The worldwide leaf economics spectrum, Nature, 428, 821–827, 2004.
Xu, X., Thornton, P. E., and Post, W. M.: A global analysis of soil microbial biomass carbon, nitrogen and phosphorus in terrestrial ecosystems, Global Ecol. Biogeogr., 22, 737–749, 2013.
Xu, Z., Yu, G., Zhang, X., Ge, J., He, N., Wang, Q., and Wang, D.: The variations in soil microbial communities, enzyme activities and their relationships with soil organic matter decomposition along the northern slope of Changbai Mountain, Appl. Soil Ecol., 86, 19–29, 2015.
Xu, Z., Yu, G., Zhang, X., He, N., Wang, Q., Wang, S., Wang, R., Zhao, N., Jia, Y., and Wang, C.: Soil enzyme activity and stoichiometry in forest ecosystems along the North-South Transect in eastern China (NSTEC), Soil Biol. Biochem., 104, 152–163, 2017.
Xu, Z., Yu, G., Zhang, X., He, N., Wang, Q., Wang, S., Xu, X., Wang, R., and Zhao, N.: Biogeographical patterns of soil microbial community as influenced by soil characteristics and climate across Chinese forest biomes, Appl. Soil Ecol., 124, 298–305, 2018.
Yu, G., Zheng, Z., Wang, Q., Fu, Y., Zhuang, J., Sun, X., and Wang, Y.: Spatiotemporal pattern of soil respiration of terrestrial ecosystems in China: the development of a geostatistical model and its simulation, Environ. Sci. Technol., 44, 6074–6080, 2010.
Zak, J. C., Willig, M. R., Moorhead, D. L., and Wildman, H. G.: Functional diversity of microbial communities: a quantitative approach, Soil Biol. Biochem., 26, 1101–1108, 1994.
Zhang, X. S. and Yang, D. A.: Allocation and study on global change transects in China, Quaternary Sci., 1, 43–52, 1995 (in Chinese).
Zhou, J., Deng, Y., Shen, L., Wen, C., Yan, Q., Ning, D., Qin, Y., Xue, K., Wu, L., He, Z., Voordeckers, J. W., Nostrand, J. D., Buzzard, V., Michaletz, S. T., Enquist, B. J., Weiser, M. D., Kaspari, M., Waide, R., Yang, Y., and Brown, J. H.: Temperature mediates continental-scale diversity of microbes in forest soils, Nat. Commun., 7, 12083, 2016.